Code
#pacman::p_load' ensures all specified packages are installed and loaded
pacman::p_load(tmap, sf, DT, stplanr, performance,
ggpubr, tidyverse)Muhamad Ameer Noor
December 1, 2023
December 14, 2023
Spatial interaction is the movement of people, goods, or information between different places. It includes things like shipping goods, energy transfer, global trade of rare items, as well as flight schedules, traffic during busy times, and people walking around.
Imagine each movement as a specific trip from one place to another. We can represent these trips using a table where one side shows where the journey starts, and the other side shows where it ends. This table is called an origin/destination matrix or a spatial interaction matrix.
In this practical exercise, we will learn to create this matrix using data about how many passengers travel between different bus stops. The data is from LTA DataMall
Learning Objectives:
Import and extract Origin-Destination (OD) data for a specific time period.
Import and store geospatial data, such as bus stops and MRT (MPSZ), as sf tibble data frame objects.
Assign planning subzone codes to bus stops in the sf tibble data frame.
Create geospatial data representing desire lines based on the OD data.
Visualize passenger volume between origin and destination bus stops using the desire lines data.
We’ll employ various R packages which will be loading using pacman on the following code
tmap: A comprehensive package for creating thematic maps that are static, interactive, or animated, specializing in visualizing geospatial data.
sf: An R package that provides simple features access for handling and manipulating geospatial data, enabling easy and straightforward operations on geographic data.
DT: This package is an R interface to the DataTables library, and it allows for the creation of interactive tables in R Markdown documents and Shiny applications.
stplanr: Designed for sustainable transport planning with R, this package assists in working with spatial data on transport systems, including the creation of desire lines, route networks, and more.
performance: This package is used for checking the performance of statistical models, including diagnostics for regression models, making it easier to assess model quality and fit.
ggpubr: Provides a convenient interface to ggplot2, especially for creating publication-ready plots with minimal amounts of code adjustments.
tidyverse: A collection of R packages designed for data science that makes it easier to import, tidy, transform, visualize, and model data.
We begin by importing the Passenger Volume by Origin Destination Bus Stops:
# The dataset is converted to a dataframe with appropriate data types
odbus202308 <- read_csv("../data/aspatial/origin_destination_bus_202308.csv.gz")
odbus202308 <- data.frame(lapply(odbus202308, factor))
odbus202308$TOTAL_TRIPS <- as.numeric(odbus202308$TOTAL_TRIPS)
odbus202308$TIME_PER_HOUR <- as.numeric(odbus202308$TIME_PER_HOUR)
# display the summary of the dataset
glimpse(odbus202308)Rows: 5,709,512
Columns: 7
$ YEAR_MONTH <fct> 2023-08, 2023-08, 2023-08, 2023-08, 2023-08, 2023-…
$ DAY_TYPE <fct> WEEKDAY, WEEKENDS/HOLIDAY, WEEKENDS/HOLIDAY, WEEKD…
$ TIME_PER_HOUR <dbl> 17, 17, 15, 15, 18, 18, 18, 18, 8, 18, 15, 11, 11,…
$ PT_TYPE <fct> BUS, BUS, BUS, BUS, BUS, BUS, BUS, BUS, BUS, BUS, …
$ ORIGIN_PT_CODE <fct> 04168, 04168, 80119, 80119, 44069, 44069, 20281, 2…
$ DESTINATION_PT_CODE <fct> 10051, 10051, 90079, 90079, 17229, 17229, 20141, 2…
$ TOTAL_TRIPS <dbl> 7, 2, 3, 10, 5, 4, 3, 22, 3, 3, 7, 1, 3, 1, 3, 1, …lapply) into a data frame.factor function to each column in the data frame, converting them to factors.For this exercise, our focus is on weekday commuting flows between 6 and 9 AM.
# Filtering and aggregating data for specific time and day
odbus6_9 <- odbus202308 %>%
filter(DAY_TYPE == "WEEKDAY") %>%
filter(TIME_PER_HOUR >= 6 & TIME_PER_HOUR <= 9) %>%
group_by(ORIGIN_PT_CODE, DESTINATION_PT_CODE) %>%
summarise(TRIPS = sum(TOTAL_TRIPS))
# save the output in rds format
write_rds(odbus6_9, "../data/rds/odbus6_9.rds")
# code for re-importing the rds file (for future use)
odbus6_9 <- read_rds("../data/rds/odbus6_9.rds")
# Displaying the data in an interactive table format
datatable(odbus6_9)readRDS) from base R re-imports an R object from an RDS file.The RDS format in R is a specialized file format used for storing single R objects. It’s a compact binary format that preserves the exact structure of the saved object, including metadata. This format is particularly efficient for saving and loading objects in R, as it ensures that the object is restored exactly as it was when saved, without any need for reformatting or reassembling data.
Using the RDS format is beneficial because it allows for fast and efficient storage and retrieval of R objects, making it ideal for situations where you need to save an object and reload it later in another session without any loss of information. The functions write_rds and read_rds (or writeRDS and readRDS in base R) are used for saving to and reading from this format, respectively. RDS is especially useful for large datasets or complex objects where preservation of structure is crucial.
We’ll use two geospatial datasets:
The datasets will be imported as follows:
Reading layer `BusStop' from data source `C:\ameernoor\ISSS624\data\geospatial' using driver `ESRI Shapefile'
Simple feature collection with 5159 features and 3 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 3970.122 ymin: 26482.1 xmax: 48280.78 ymax: 52983.82
Projected CRS: SVY21Reading layer `MPSZ-2019' from data source `C:\ameernoor\ISSS624\data\geospatial' using driver `ESRI Shapefile'
Simple feature collection with 332 features and 6 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 103.6057 ymin: 1.158699 xmax: 104.0885 ymax: 1.470775
Geodetic CRS: WGS 84Simple feature collection with 332 features and 6 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 2667.538 ymin: 15748.72 xmax: 56396.44 ymax: 50256.33
Projected CRS: SVY21 / Singapore TM
First 10 features:
SUBZONE_N SUBZONE_C PLN_AREA_N PLN_AREA_C REGION_N
1 MARINA EAST MESZ01 MARINA EAST ME CENTRAL REGION
2 INSTITUTION HILL RVSZ05 RIVER VALLEY RV CENTRAL REGION
3 ROBERTSON QUAY SRSZ01 SINGAPORE RIVER SR CENTRAL REGION
4 JURONG ISLAND AND BUKOM WISZ01 WESTERN ISLANDS WI WEST REGION
5 FORT CANNING MUSZ02 MUSEUM MU CENTRAL REGION
6 MARINA EAST (MP) MPSZ05 MARINE PARADE MP CENTRAL REGION
7 SUDONG WISZ03 WESTERN ISLANDS WI WEST REGION
8 SEMAKAU WISZ02 WESTERN ISLANDS WI WEST REGION
9 SOUTHERN GROUP SISZ02 SOUTHERN ISLANDS SI CENTRAL REGION
10 SENTOSA SISZ01 SOUTHERN ISLANDS SI CENTRAL REGION
REGION_C geometry
1 CR MULTIPOLYGON (((33222.98 29...
2 CR MULTIPOLYGON (((28481.45 30...
3 CR MULTIPOLYGON (((28087.34 30...
4 WR MULTIPOLYGON (((14557.7 304...
5 CR MULTIPOLYGON (((29542.53 31...
6 CR MULTIPOLYGON (((35279.55 30...
7 WR MULTIPOLYGON (((15772.59 21...
8 WR MULTIPOLYGON (((19843.41 21...
9 CR MULTIPOLYGON (((30870.53 22...
10 CR MULTIPOLYGON (((26879.04 26...dsn) and layer.mpsz data) to a file in RDS format.We’ll merge the planning subzone codes from the mpsz dataset into the busstop dataset.
busstop and mpsz. It returns the areas that are common to both spatial objects.BUS_STOP_N, SUBZONE_C) in the resulting data frame.Now, let’s append the planning subzone codes to the odbus6_9 dataset:
Rows: 227,098
Columns: 4
Groups: ORIGIN_BS [5,005]
$ ORIGIN_BS <chr> "01012", "01012", "01012", "01012", "01012", "01012", "01012…
$ DESTIN_BS <fct> 01112, 01113, 01121, 01211, 01311, 07371, 60011, 60021, 6003…
$ TRIPS <dbl> 177, 111, 40, 87, 184, 18, 22, 16, 12, 21, 2, 3, 1, 1, 1, 1,…
$ ORIGIN_SZ <chr> "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", …left_join function specifically keeps all rows from the left data frame and adds matching rows from the right data frame. If there is no match, the right side will contain NA.ORIGIN_PT_CODE to ORIGIN_BS, SUBZONE_C to ORIGIN_SZ, and DESTINATION_PT_CODE to DESTIN_BS.Checking for duplicate records is crucial, if duplicates exist, we keep only unique records:
Rows: 226,610
Columns: 4
Groups: ORIGIN_BS [5,005]
$ ORIGIN_BS <chr> "01012", "01012", "01012", "01012", "01012", "01012", "01012…
$ DESTIN_BS <fct> 01112, 01113, 01121, 01211, 01311, 07371, 60011, 60021, 6003…
$ TRIPS <dbl> 177, 111, 40, 87, 184, 18, 22, 16, 12, 21, 2, 3, 1, 1, 1, 1,…
$ ORIGIN_SZ <chr> "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", …We’ll now complete the dataset with destination subzone codes:
Rows: 227,458
Columns: 5
Groups: ORIGIN_BS [5,005]
$ ORIGIN_BS <chr> "01012", "01012", "01012", "01012", "01012", "01012", "01012…
$ DESTIN_BS <chr> "01112", "01113", "01121", "01211", "01311", "07371", "60011…
$ TRIPS <dbl> 177, 111, 40, 87, 184, 18, 22, 16, 12, 21, 2, 3, 1, 1, 1, 1,…
$ ORIGIN_SZ <chr> "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", …
$ DESTIN_SZ <chr> "RCSZ10", "DTSZ01", "RCSZ04", "KLSZ09", "KLSZ06", "KLSZ06", …left_join function specifically keeps all rows from the left data frame and adds matching rows from the right data frame. If there is no match, the right side will contain NA.ORIGIN_PT_CODE to ORIGIN_BS, SUBZONE_C to ORIGIN_SZ, and DESTINATION_PT_CODE to DESTIN_BS.Checking for and removing any remaining duplicates:
Rows: 226,846
Columns: 5
Groups: ORIGIN_BS [5,005]
$ ORIGIN_BS <chr> "01012", "01012", "01012", "01012", "01012", "01012", "01012…
$ DESTIN_BS <chr> "01112", "01113", "01121", "01211", "01311", "07371", "60011…
$ TRIPS <dbl> 177, 111, 40, 87, 184, 18, 22, 16, 12, 21, 2, 3, 1, 1, 1, 1,…
$ ORIGIN_SZ <chr> "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", "RCSZ10", …
$ DESTIN_SZ <chr> "RCSZ10", "DTSZ01", "RCSZ04", "KLSZ09", "KLSZ06", "KLSZ06", …Finally, we’ll prepare the data for visualisation:
Rows: 20,595
Columns: 3
Groups: ORIGIN_SZ [309]
$ ORIGIN_SZ <chr> "AMSZ01", "AMSZ01", "AMSZ01", "AMSZ01", "AMSZ01", "AMSZ01…
$ DESTIN_SZ <chr> "AMSZ01", "AMSZ02", "AMSZ03", "AMSZ04", "AMSZ05", "AMSZ06…
$ MORNING_PEAK <dbl> 1866, 8726, 12598, 2098, 7718, 1631, 1308, 2261, 1526, 14…NA) in the data frame.ORIGIN_SZ and DESTIN_SZ.summarize) from dplyr package calculates summary statistics for each group, here summing up the TRIPS for each ORIGIN_SZ and DESTIN_SZ combination.We’ll now create and visualize desire lines using the stplanr package.
Intra-zonal flows are not required for our analysis:
Rows: 20,304
Columns: 3
Groups: ORIGIN_SZ [309]
$ ORIGIN_SZ <chr> "AMSZ01", "AMSZ01", "AMSZ01", "AMSZ01", "AMSZ01", "AMSZ01…
$ DESTIN_SZ <chr> "AMSZ02", "AMSZ03", "AMSZ04", "AMSZ05", "AMSZ06", "AMSZ07…
$ MORNING_PEAK <dbl> 8726, 12598, 2098, 7718, 1631, 1308, 2261, 1526, 141, 655…Here’s how to create desire lines using the od2line() function:
In the context of transport planning, desire lines are like the paths people would naturally prefer to take when going from one place to another. Think of them as the routes you would choose if you could walk or travel in a straight line, without any obstacles.
Imagine a park where there’s a paved path, but people consistently walk across the grass to get from one side to the other. The worn-down grassy trail is the desire line – it shows where people naturally want to walk, even if it’s not the officially designated path.
In transport planning, understanding these desire lines helps city planners decide where to put roads, sidewalks, or public transportation routes. It’s about making sure the infrastructure fits the way people actually move around, making travel more efficient and convenient for everyone.
Rows: 20,304
Columns: 4
$ ORIGIN_SZ <chr> "AMSZ01", "AMSZ01", "AMSZ01", "AMSZ01", "AMSZ01", "AMSZ01…
$ DESTIN_SZ <chr> "AMSZ02", "AMSZ03", "AMSZ04", "AMSZ05", "AMSZ06", "AMSZ07…
$ MORNING_PEAK <dbl> 8726, 12598, 2098, 7718, 1631, 1308, 2261, 1526, 141, 655…
$ geometry <LINESTRING [m]> LINESTRING (29501.77 39419...., LINESTRING (29…Without any filtering, the desire lines are quite messy with various flows entangled.
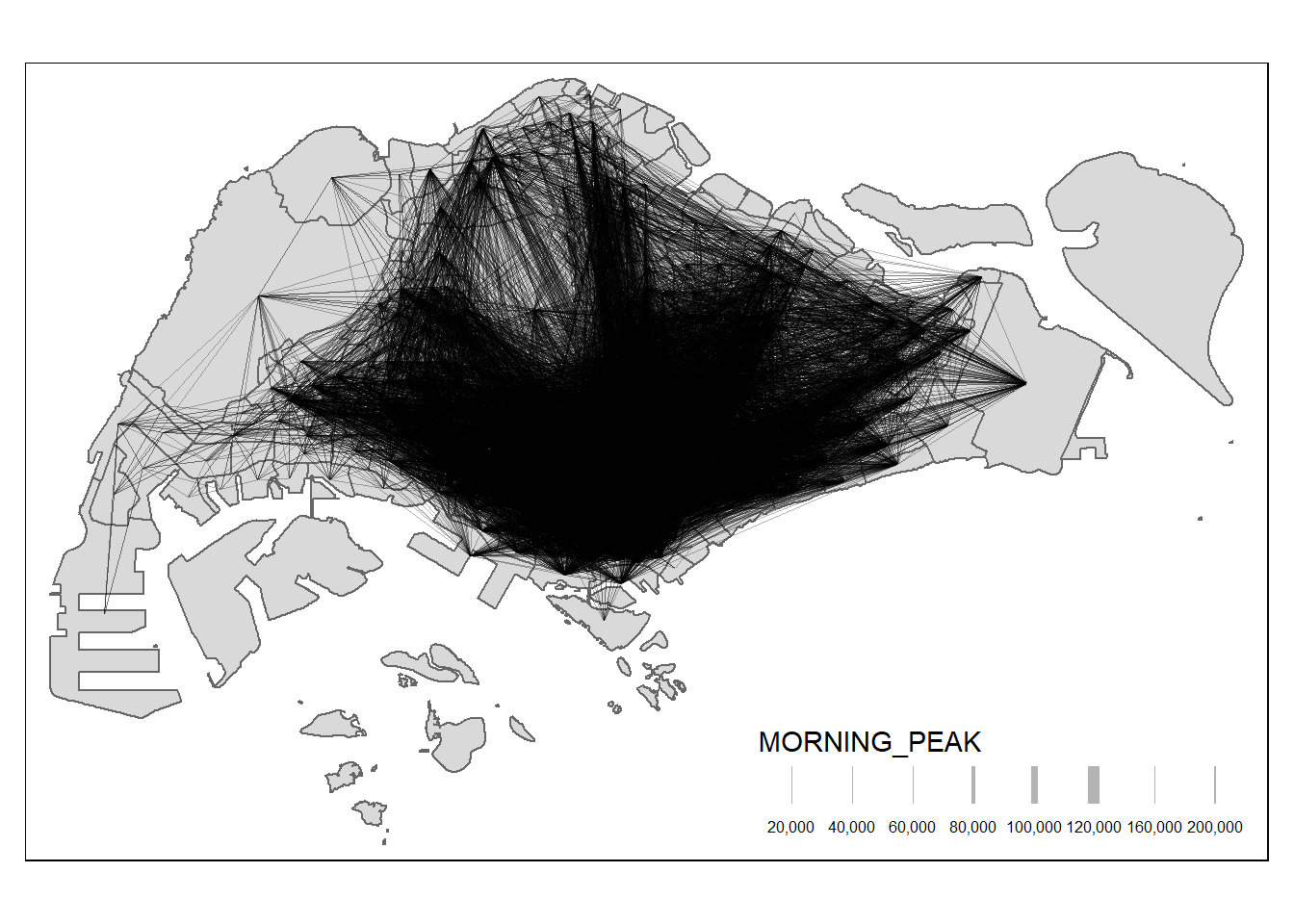
mpsz data.MORNING_PEAK, with the style defined as “quantile” and specific scaling parameters.Focusing on selected flows can be insightful, especially when data are skewed:
MORNING_PEAK is greater than or equal to 5000.MORNING_PEAK variable. The style is set to “quantile”, with a specific scale and number of breaks (n) for line width, and the color is set to orange.MORNING_PEAK.